Abstract
Introduction
Despite recent advances in therapy, acute myeloid leukemia (AML) remain a medical challenge with high morbidity and mortality rates. For most patients, allogeneic hematopoietic stem cell transplantation remain the only curative option, but due to the advanced age at diagnosis, a significant proportion of patients are not elegible to this form of therapy. Nevertheless, novel therapies are warranted. There is preclinical evidence that anti-inflammatory compounds, such as COX-2 inhibitors and steroids, may have anti-neoplastic activity in different tumor types, including AML; nevertheles the mechanisms associated with its anti-neoplastic activity are not clear. Therefore, the aim of this work was to evaluate the anti-leukemic effect of the anti-inflammatory compounds nimesulide and prednisolone in AML cell lines and to identify genes and molecular pathways associated with cytotoxicity through transcriptome analysis.
Methods
The leukemic cell lines HL-60, THP-1, OCI-AML2 and OCI-AML3 were treated with nimesulide and prednisolone at 100 µM alone and in combination and with cytarabine at 2.5 µM. Twenty four hours after treatment , we measured the amount of cell death using Annexin V Apoptosis Detection Kit FITC (ThermoFisher) and the cell cycle was analized after fixing the cells with 70% alcohol and incubation with propidium iodide (1mg/ml) and RNAse (10mg/ml). In another experiment, we harvested the cells after 4 hours of treatment for transcriptome analysis. RNA was extracted from control (DMSO) and treatment groups (1 - nimesulide, 2 - prednisolone , 3 - nimesulide and prednisolone) with RNeasy Mini Kit (Qiagen). The Illumina® NEBNext® Ultra II Directional RNA Library Prep Kit was used for library preparation, following the manufacturer protocol using Poly(A) mRNA Magnetic Isolation Module. Equimolar amount of libraries was sequenced using an Illumina NextSeq 500, following the manufacturer's instructions, on the Oklahoma Medical Research Foundation Genomics Core (USA). The sequences obtained with the RNA-Seq technique were aligned in the human genome of reference GRCh37.75 by the software Spliced Transcripts Alignment to a Reference (STAR) v2.5 and to obtain normalized counts in FPKM, the software Expectation-Maximization (RSEM) v1.3.0 was used. To identify network of genes correlated with the treatment (modules), we used the Weighted Correlation Network Analysis (WGCNA). Functional enrichment analysis of the WGCNA differentially expressed modules was performed using the Integrated Annotation, Visualization and Discovery Database (DAVID) v6.8 in order to correlate with biological processes.
Results
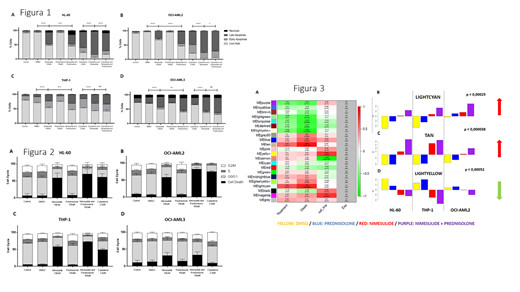
In the cell cycle analysis, we observed a significant increase (p < 0.05) in the sub-G0 phase (cell death) after treatment with nimesulide alone, and in combination with prednisolone (figure 1). No effect was observed in the prednisolone only group. The cell cycle effect induced by nimesulide on HL-60 and OCI-AML2 was similar to the induced by cytarabine, a standard chemotherapy agent for AML that in known to induce arrest in the S phase. In addition, the cell line arrest in THP-1 was greater with nimesulide than with cytarabine, while OCI-AML3 was less sensitive to both nimesulide and cytarabine. Regarding cell death mechanism, treatment with nimesulide induced predominantly an increase in late apoptosis that was potentiated after combined treatment with nimesulide and cytarabine (figure 2). After the demonstration of cell cycle arrest and apoptosis induction after treatment with nimesulide, we performed whole transcriptome sequencing followed by WGCNA analysis. We have identified gene modules that were significantly correlated with anti-inflammatory treatments, being 1 module down-regulated (lightyellow with p = 0.00052) and 2 modules up-regulated (lightcyan with p = 0 .00025 and tan with p = 0.000038). Analysis of functional enrichment using DAVID showed up-regulation of gene networks associated with apoptotic processes and autophagy and down- regulation of gene networks associated with cell cycle and RNA splicing pathways
Conclusions
The COX-2 inhibitor nimesulide caused cell cycle arrest and apoptosis in AML cell lines and potentiated the cytotoxic effects of cytarabine. This treatment was associated with up- regulation of autophagy and apoptosis and down-regulation of cell cycle and RNA splicing gene networks.
Santos: Novartis: Membership on an entity's Board of Directors or advisory committees, Research Funding; Amgen: Membership on an entity's Board of Directors or advisory committees; Abbvie: Membership on an entity's Board of Directors or advisory committees; Pfizer: Consultancy, Membership on an entity's Board of Directors or advisory committees. Campregher: Astellas: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal